What is the MMHub?
The Mulberry Metabolome Database (MMHub) is a comprehensively, freely, and user-friendly available electronic database containing detailed information (structure, MS data, and so on) about all metabolites identified in the mulberry species. It is intended to be used for applications in metabolomics, traditional medicines, biomarker discovery and general education.
Now, the database (version 1.0) contains 124 metabolites and 812 metabolite features of mulberry species.
Workflow of metabolite profiling and related data analysis in mulberry species.
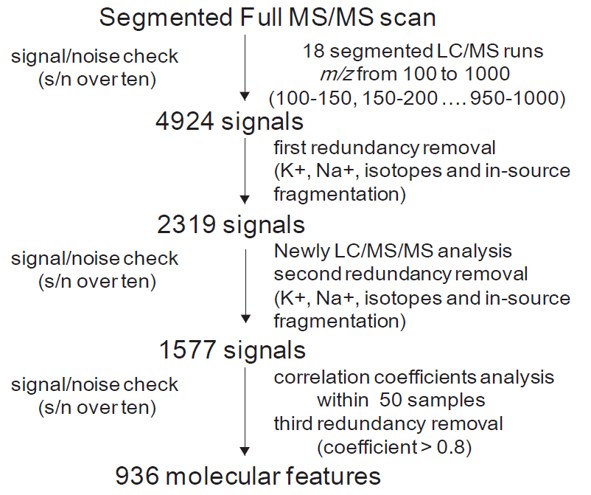
A widely-targeted metabolomics approach based on high-performance liquid chromatography mass spectrometer (HPLC-MS/MS) was carried out in this study. By application of a segmented Full-MS/MS scan strategy, metabolites with mass range from m/z 100 to 1,000 were high-sensitivity detected in 18 LC/MS/MS runs. About 4900 chromatographic peaks with signal/noise (s/n) >10 were manually checked out (data not shown). To produce a matrix containing fewer biased and redundant data, redundancies that caused by isotopes, in-source fragmentation, K+, Na+, and NH4+ adduct, and dimerization was removed by using an in-house-developed software written in Perl. In addition, we believed that transitions derived from the same metabolites would have high-correlation among different samples. Thus, a pre-experiment contains 50 mulberry varieties were then carried out to figure out the potential redundancy. More than 650 metabolites with high-correlation coefficients (over 0.8) and the same retention time (± 0.1min) were considered as redundancy. Finally, a MS2 spectral tag (MS2T) library contains 936 distinct metabolite features (almost non-redundant) were obtained and reported as recommended previously. The data matrix was then used for metabolite identification/annotation and semi-quantification.
Metabolite identification/annotation was based on the accurate m/z, the retention time (RT) and the fragmentation patterns delivered by a Q Exactive LC-MS/MS system. In this study, 35 commercially available standards were purchased and identified using the same profiling conditions as the extracts, including amino acids, flavonoids, alkaloids, and vitamins. Eighty-eight metabolite features were putatively identified by query the MS/MS spectral data taken from the literature or to search the databases (MassBank, KNApSAcK, HMDB, and METLIN). It is noteworthy that 35.5% (44/123) of these metabolites features were classified as flavonoids, which was identified based on the fragmentation pattern (delivered by MS/MS spectra). For instance, m1903 (RT 7.35min, m/z 595.1652, error -0.84 ppm) and m1963 (RT 6.81min, m/z 611.1590, error -2.78 ppm) were identified as kaempferol O-rhamnosyl-O-hexoside (m/z 287.0539, 153.0179, and 121.0282 are the diagnostic fragment ions of Kaempferol, the natural loss of hexoside (162 Da, 449.1071 → 287.0539) and rhamnoside (146 Da, 595.1650 → 449.1071) are revealed in the mass spectrum) and quercetin O-rhamnosyl-O-hexoside (m/z 303.0494, 257.0434, 229.0491, and 153.0180 are the diagnostic fragment ions of Quercetin). Based on this strategy, the data matrix with 124 annotated metabolites has been constructed and used for the following studies.
Metabolite Reporting Checklist
| Aspect | Information | Fill in |
| Type of metabolome analysis | targeted metabolite analysis | FALSE |
| non-targeted metabolite class scale profiling | FALSE | |
| non-targeted metabolome scale profiling | TRUE | |
| non-targeted finger printing of mass features | FALSE | |
| Type of quantification | absolute or quantification | Quantification |
| Type of reference samples | chemcially defined | FALSE |
| biologically defined | More than 50 divergent samples were mixed as the reference sample | |
| Type of replication | analytical (same analytical sample preparation) | 1 |
| technological (same biological preparation) | 0 | |
| biological (same experimental condition) | 2 | |
| full experiment | 2 | |
| Type of technology | reference publication | Plant Cell reference style |
| Sample preparation | method of sample preparation | Chemically non-modified|derivatized (70% methanol) |
| method of chromatography/separation | ||
| method of ionization | ESI | |
| method of detection | specify detection type (MS) and instrumentation (Thermo Scientific™ Q Exactive™ hybrid quadrupole-Orbitrap mass spectrometer) | |
| Metabolite | metabolite name | m**** |
| metabolite sum formula | The formula were calculated by Qualitative Analysis B.04.00 | |
| metabolite structure and public source of metabolite identifier | Massbank_database、METLIN-Scripps Center for Metabolomics and Spectrometry and Human Metabolome Database | |
| Identification | identification process | manually supervized |
| by authentic mass isotopomer added to one or all biological sample(s) | FALSE | |
| by authentic reference compound within a co-processed reference mixture | TRUE | |
| by authentic reference compound previously mapped to the analytical system | TRUE | |
| reference library | Massbank_database、METLIN-Scripps Center for Metabolomics and Spectrometry and Human Metabolome Database | |
| type of mass spectrum | Thermo Scientific™ Q Exactive™ hybrid quadrupole-Orbitrap mass spectrometer | |
| by match of molecular mass (single mass fragment) | The molecular mass were correct to 0.0001 | |
| by match of fragments | Match the main fragments | |
| by match of fragmentation pattern | Specify set of mass fragment | |
| by match of mass spectrum to reference library | Match the molecular mass and the main fragments | |
| type of retention index | FALSE | |
| by match of retention time (index) to reference library | FALSE | |
| Quantification | type of quantification | Relative quantification |
| Validity testing | Recovery testing (chemical analog) | FALSE |
| Recovery testing (internally added mass isotopomer) | FALSE | |
| Recovery testing (mixture of most divergent samples from the experiment) | FALSE | |
| Test for linear range | FALSE | |
| Limit of quantification (LOQ) | FALSE | |
| Limit of detection (LOD) | FALSE |
Mulberry Resources
| ID | Name |
|---|---|
| ml001 | M. alba var. Taiwansijiguosang |
| ml002 | M. alba var. Taiwanlongguosang |
| ml003 | M. alba var. Taiwandaguosang |
| ml004 | M. alba var. Baiyuwang |
| ml005 | M. alba var. 8632 |
| ml006 | M. alba var. Wittiorum |
| ml007 | M. alba var. Jinqiang No.82 |
| ml008 | M. alba var. Jinqiang No.63 |
| ml009 | M. alba var. Gui 23 |
| ml010 | M. alba var. Emeihuasang |
| ml011 | M. alba var. Ichibei 34 |
| ml012 | M. alba var. Langzhongdayesang |
| ml013 | M. alba var. 603133 |
| ml014 | M. alba var. Yusang |
| ml015 | M. alba var. Banqiao No.6 |
| ml016 | M. alba var. 607138 |
| ml017 | M. alba var. Huai 30-2 |
| ml018 | M. alba var. Lesangdahongpi 30 |
| ml019 | M. alba var. Tuosang 25 |
| ml020 | M. alba var. Heyebai 36 |
| ml021 | M. alba var. Jiadingsang 29 |
| ml022 | M. alba var. Yidashoumusang 31 |
| ml023 | M. alba var. Wupisang |
| ml024 | M. alba var. Jimengsang |
| ml025 | M. alba var. Shidian No.6 |
| ml026 | M. alba var. Dejiang No.13 |
| ml027 | M. alba var. Gui 14 |
| ml028 | M. alba var. Huosang |
| ml029 | M. alba var. Baojing No.7 |
| ml030 | M. alba var. Pisang No.2 |
| ml031 | M. alba var. Mangshi No.10 |
| ml032 | M. alba var. Tortuosa |
| ml033 | M. alba var. Shanxitiansang |
| ml034 | M. alba var. Shenglidaye |
| ml035 | M. alba var. Xinjiansang No.4 |
| ml036 | M. alba var. Lizhuang 5 |
| ml037 | M. alba var. Changyajingsang 17 |
| ml038 | M. alba var. Dayezaoshengsang |
| ml039 | M. alba var. Nongsang 14 |
| ml040 | M. alba var. Xiang No.0 |
| ml041 | M. alba var. Yun 3 |
| ml042 | M. alba var. Shin-Ichinose |
| ml043 | M. alba var. Huanglusang 13 |
| ml044 | M. alba var. Pendula 1 |
| ml047 | M. alba var. Shansang 305 |
| ml050 | M. alba var. Laevigata |
| ml051 | M. alba var. Hanguodabaizhengzhu |
| ml052 | M. alba var. Hongguo No.2 |
| ml053 | M. alba var. Kanwa |
| ml054 | M. alba var. Yun No.3 |
| ml055 | M. alba var. Shin-ichibei |
| ml056 | M. alba var. Shiping |
| ml057 | M. alba var. 6071 |
| ml058 | M. alba var. Ribengailiangshizi |
| ml060 | M. alba var. Gongnongjingsang |
| ml061 | M. alba var. 6031 |
| ml062 | M. alba var. Cathayana |
| ml063 | M. alba var. Shengnan |
| ml064 | M. alba var. Ichibei |
| ml065 | M. alba var. Shimiansang |
| ml066 | M. alba var. Yidashoumusang |
| ml067 | M. alba var. Zhuangelou |
| ml068 | M. alba var. Kuisang |
| ml069 | M. alba var. Nongsang No.8 |
| ml070 | M. alba var. Gailiangshufan |
| ml071 | M. alba var. Shanxitiansang |
| ml072 | M. alba var. Baokan 61 |
| ml073 | M. nigra |
| ml076 | M. alba var. Hongguo No.1 |
| ml079 | M. alba var. Jianpuzhai |
| ml088 | M. alba var. Pendula |
| ml089 | M. alba var. Agentingsang |
| ml091 | M. alba var. Yunguo No.1 |
| ml092 | M. alba var. Yun 1 |
| ml093 | M. alba var. Yun 2 |
| ml094 | M. alba var. Yun 3 |
| ml095 | M. alba var. Yun 4 |
| ml096 | M. alba var. Yun 5 |
| ml097 | M. alba var. Yun 6 |
| ml098 | M. alba var. Yun 7 |
| ml100 | M. alba var. Yun 9 |
| ml103 | M. alba var. Yun 12 |
| ml104 | M. alba var. Yun 13 |
| ml105 | M. alba var. Yun 14 |
| ml110 | M. alba var. Yun 19 |
| ml111 | M. alba var. Zhenzhubai |
| ml112 | M. alba var. Mongolica |
| ml114 | M. alba var. Shuangtouheyebai |
| ml115 | M. alba var. Zu 83122 |
| ml116 | M. alba var. Meiguosang |
| chuan | M. notabilis |